cellXpress 2.4.0 with WSI support available now (June 2024)
Key features
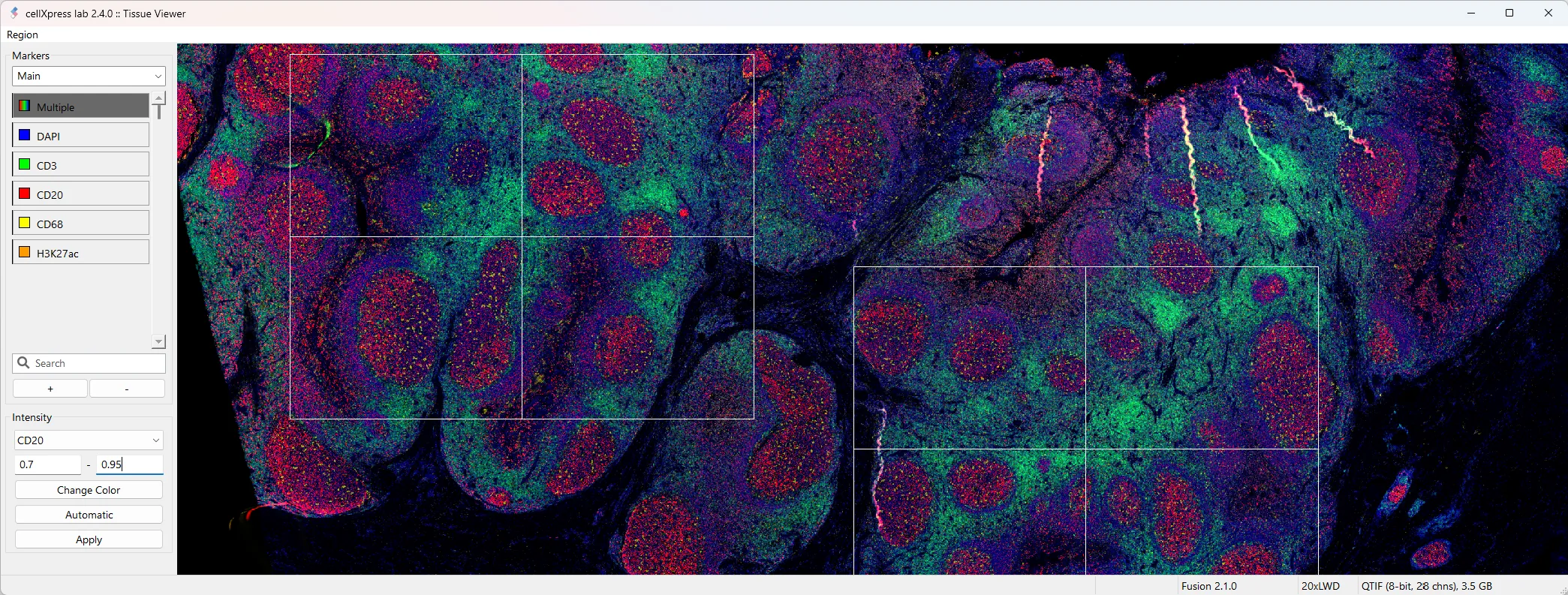
Marker-set architecture
Optimum image processing based on different marker sets
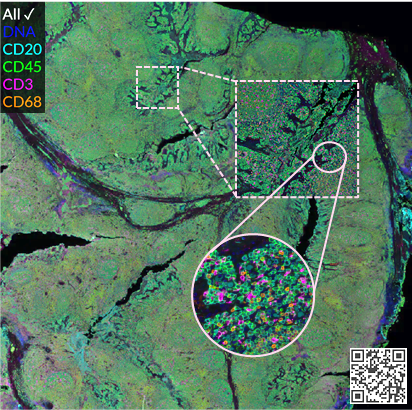
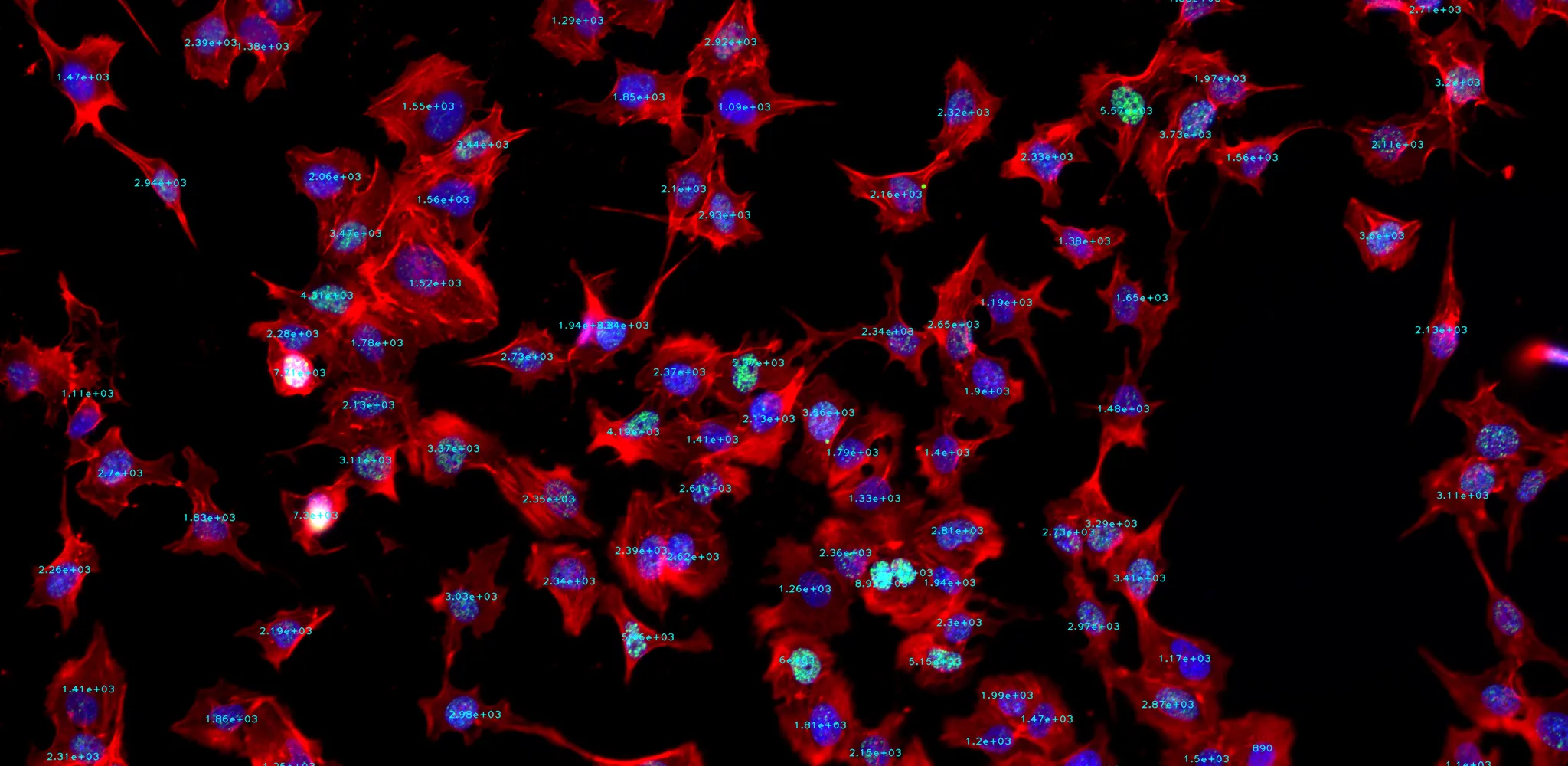
Hyperplexed images

Intuitive visualization of large tissue images and results with 50+ markers
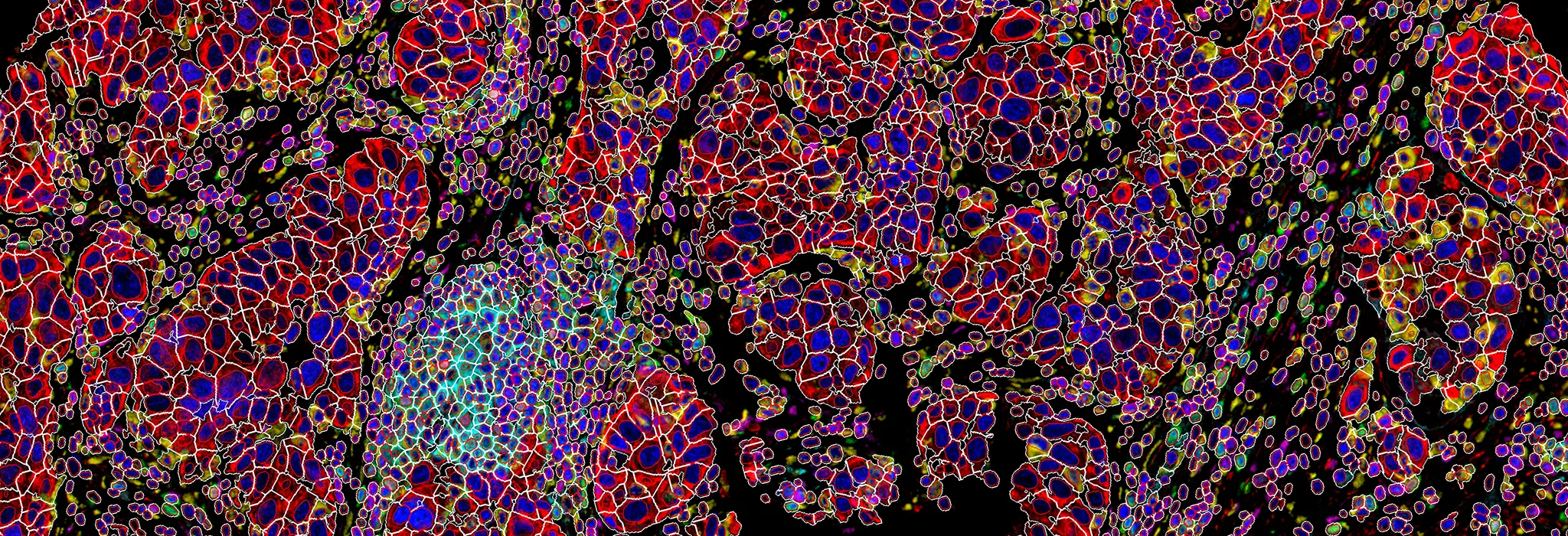
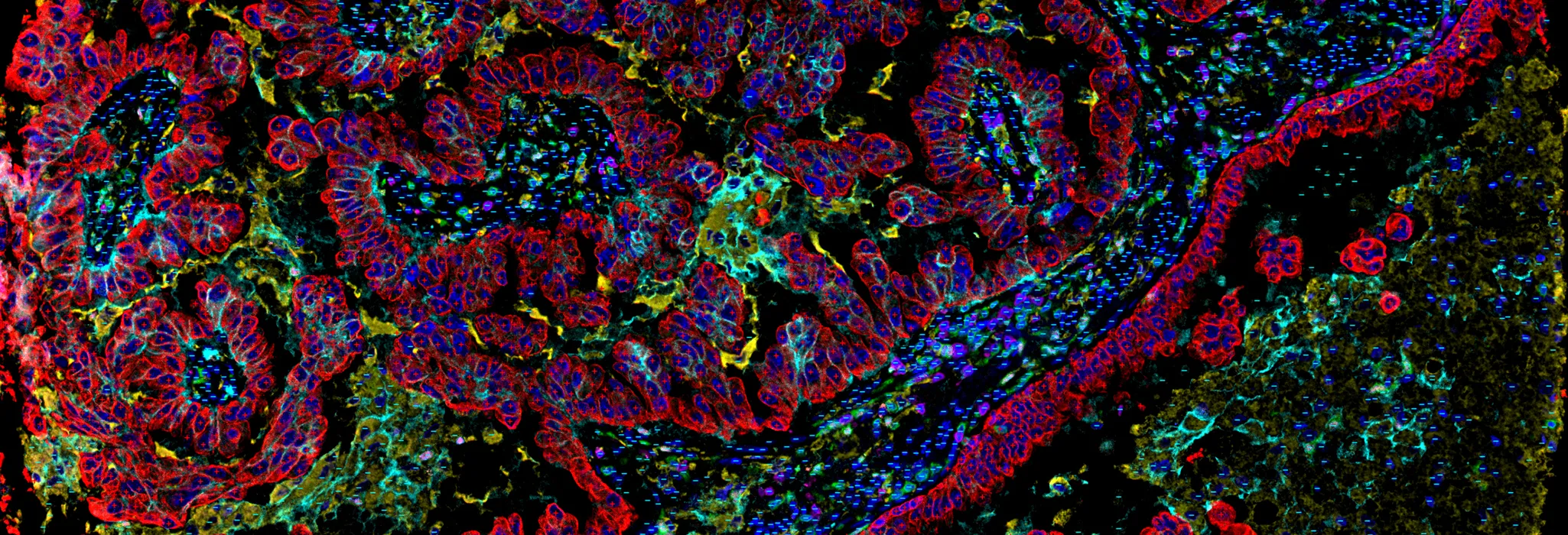
CellShape AI segmentation
Accurate detection of heterogenous and overlapped cells in tissues
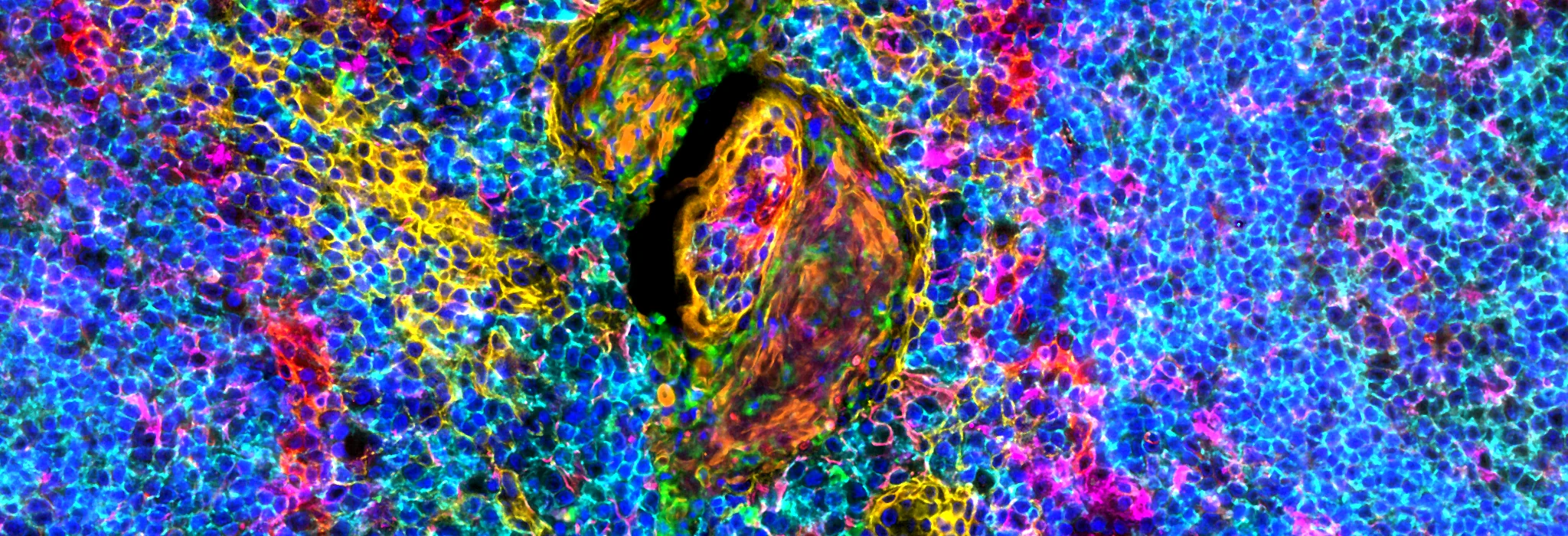
Subpopulation identification
Systematic identification of diverse cell types or subpopulations
Published work that used cellXpress
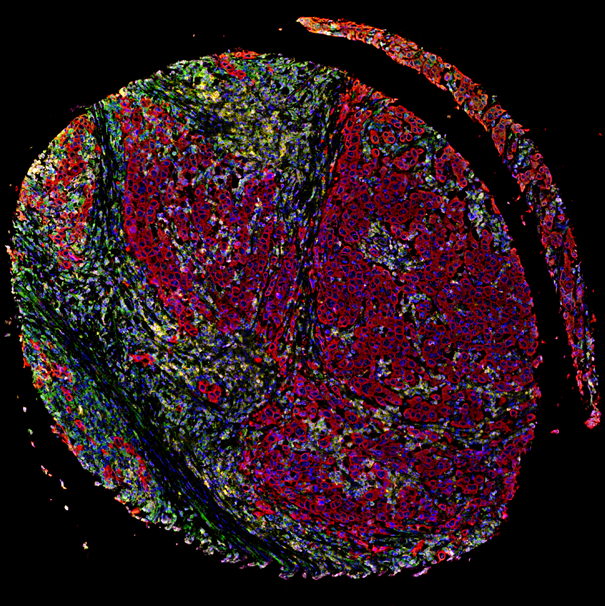
Choice of PD-L1 immunohistochemistry assay influences clinical eligibility for gastric cancer immunotherapy
J Yeong et. al., Gastric Cancer, 2022
cellXpress was used to quantify the three PD-L1 antibody clones (22C3, SP142 and 28-8) and cytokeratin.